BAD Day 1: One way ANOVA
This notebook is intended to demonstrate the application of the one way ANOVA analysis on a built in R data set: PlantGrowth
# Getting the data set and having a peek into its content
help(PlantGrowth)
PlantGrowth| weight | group |
|---|---|
| 4.17 | ctrl |
| 5.58 | ctrl |
| 5.18 | ctrl |
| 6.11 | ctrl |
| 4.50 | ctrl |
| 4.61 | ctrl |
| 5.17 | ctrl |
| 4.53 | ctrl |
| 5.33 | ctrl |
| 5.14 | ctrl |
| 4.81 | trt1 |
| 4.17 | trt1 |
| 4.41 | trt1 |
| 3.59 | trt1 |
| 5.87 | trt1 |
| 3.83 | trt1 |
| 6.03 | trt1 |
| 4.89 | trt1 |
| 4.32 | trt1 |
| 4.69 | trt1 |
| 6.31 | trt2 |
| 5.12 | trt2 |
| 5.54 | trt2 |
| 5.50 | trt2 |
| 5.37 | trt2 |
| 5.29 | trt2 |
| 4.92 | trt2 |
| 6.15 | trt2 |
| 5.80 | trt2 |
| 5.26 | trt2 |
We will now use the summary functon to get more information about the whole
data set
# Getting a basic summary of the data
summary(PlantGrowth) weight group
Min. :3.590 ctrl:10
1st Qu.:4.550 trt1:10
Median :5.155 trt2:10
Mean :5.073
3rd Qu.:5.530
Max. :6.310
You can also get statistical information on a single attribute of the data
# Computing the SD of a given attribute
sd(PlantGrowth$weight)0.701191842508168
Or per groups within the data set
# Compute the sd of each group for the given attribute
tapply(PlantGrowth$weight, PlantGrowth$group, sd)- ctrl
- 0.583091378392406
- trt1
- 0.793675696434703
- trt2
- 0.442573283322786
The next thing we will do is creating a boxplot of the data set (as in the main Tutorial).
boxplot(weight ~ group, data = PlantGrowth, col = 'mediumpurple',
xlab = 'Group', ylab = 'Weight')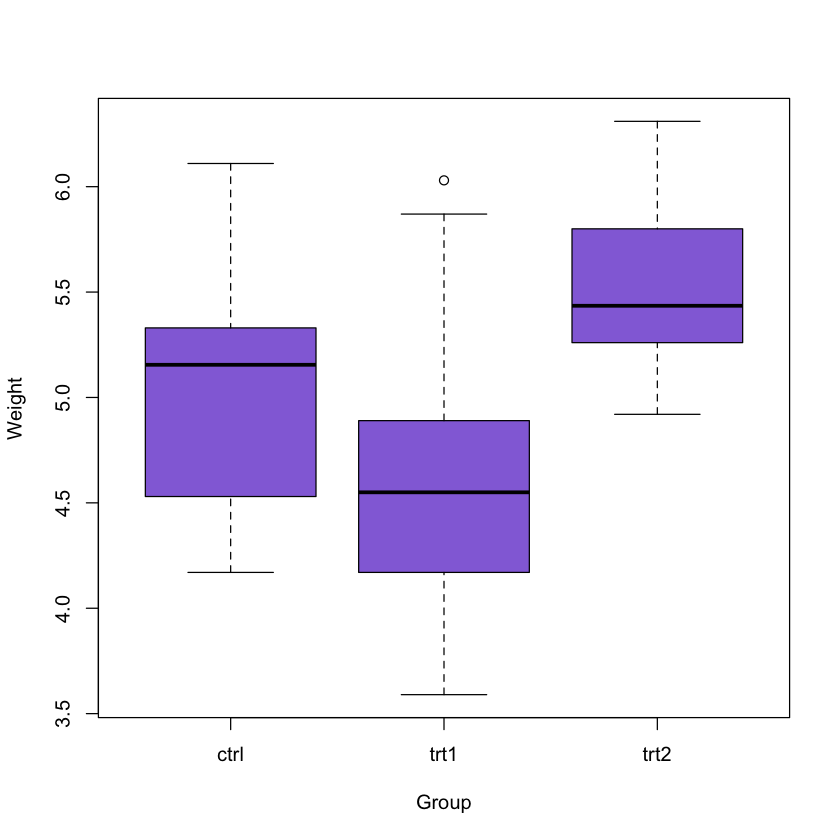
We can also create histograms of the data. In this case we will use the lattice plotting system:
# Creating a conditional histogram of the data
par(mfrow = c(1,3))
# We will use the lattice plotting system
library(lattice)
# Creating the histogram
histogram(~weight | group, data = PlantGrowth, col = 'mediumpurple', type = 'count')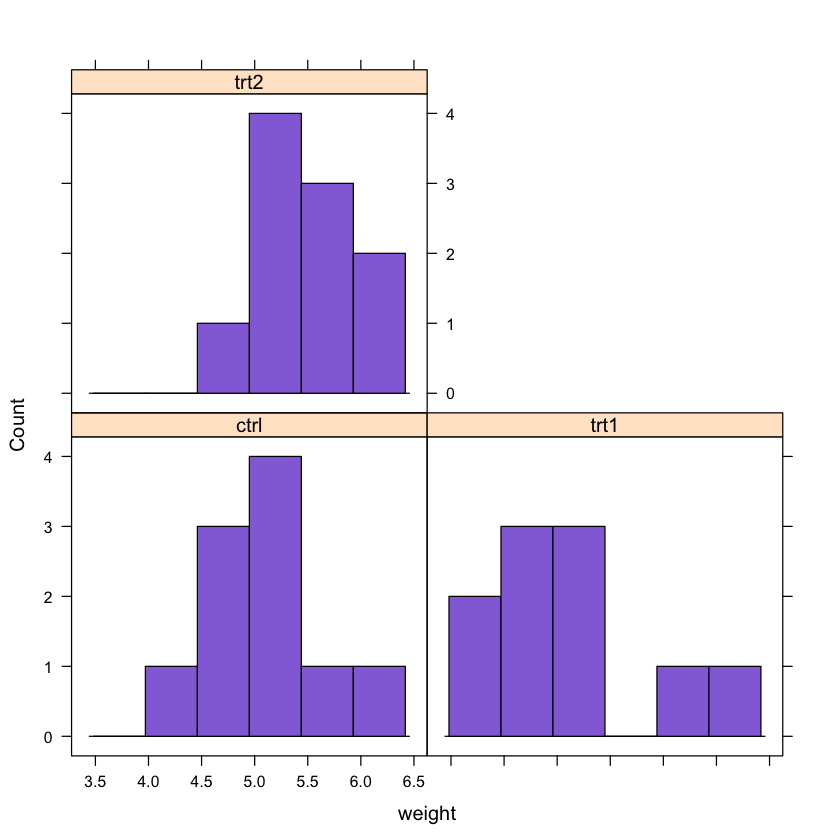
** Our null hypotesis is that the three groups have the same growth mean**
First we will use a completely randomized design one-way ANOVA
# One way ANOVA (Randomized design)
analysis <- aov(weight ~ group, data = PlantGrowth)
summary(analysis) Df Sum Sq Mean Sq F value Pr(>F)
group 2 3.766 1.8832 4.846 0.0159 *
Residuals 27 10.492 0.3886
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Now we will build a linear model with lm() and will then use the anova()
command to analyse the fit
fit <- lm(weight ~ group, data = PlantGrowth)
anova(fit)| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| group | 2 | 3.76634 | 1.8831700 | 4.846088 | 0.01590996 |
| Residuals | 27 | 10.49209 | 0.3885959 | NA | NA |
As expected, both approaches provide the same results!